5. Pipeline#
Thus far, we’ve applied the preprocessing, epoching, and averaging steps to a single participant only. However, we typically study a larger group of participants. This is because (a) data from a single participant is typically quite noisy, and (b) we want to generalize our findings to the population from which our participants were drawn.
Therefore, we need to repeat the analysis for all participants by creating an analysis pipeline that takes each participant’s raw data as an input and performs the same set of processing steps on them.
We will first do this manually (using a loop) and then with a pre-packaged automatic pipeline function.
Goals
Repeating the processing steps for all participants
Using a fully automated pipeline
5.1. Load Python packages#
The functions for our custom pipeline are again provided by the MNE-Python package (Gramfort et al, 2013) and pandas. We will also use the hu-neuro-pipeline package for the fully automated pipeline, plus two new packages (matplotlib and seaborn) for visualization.
# %pip install mne hu-neuro-pipeline pandas matplotlib seaborn
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from mne import Epochs, combine_evoked, merge_events, set_bipolar_reference
from mne.io import read_raw
from mne.preprocessing import ICA
from mne.viz import plot_compare_evokeds
from pipeline import group_pipeline
from pipeline.datasets import get_erpcore
5.2. Custom pipeline#
We’ll start by creating a custom pipeline that repeats the preprocessing, epoching, and averaging steps for all participants.
This is as easy as taking all the code from the previous chapters and, instead of reading only a single raw EEG file, putting it inside a for loop that iterates over all raw EEG files.
Let’s first download some more datasets (for the sake of time, we’re only using 10 participants instead of all 40):
files_dict = get_erpcore('N170', participants=10, path='data')
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c4eba01090877890aa3' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c50ba0109087e8916aa' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c5286541a08f914ab2d' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//610215220c4cba026dbce4d0' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//610215260c4cba0277bc7a17' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c4be80d3708caa58293' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-001/eeg/sub-001_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c42e80d3708c5a57469' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c5486541a090014bb81' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c5c86541a08f814a3b7' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c5fe80d3708caa582c7' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c61ba01090876890471' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c63e80d3708c1a56934' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102152d0c4cba026dbce4e7' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-002/eeg/sub-002_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//610215310c4cba026abcd4ee' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102153c317620028637d408' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021537317620027d389014' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c7486541a090014bbec' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c72e80d3708c7a58090' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c66e80d3708caa582de' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c6dba0109087e8916f0' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c6fba01090873890087' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102154dc7a976029b9e5bfc' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c88ba0109087a8915d5' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c9086541a08fc14b889' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c92e80d3708c7a580c6' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c9586541a08fc14b894' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c97e80d3708c3a56bdf' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-005/eeg/sub-005_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021551317620027838216b' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102155a317620028737d513' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ca7ba0109087a891613' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021556c7a976029a9e2033' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ca5ba0109087a89160b' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075c9986541a090014bc3c' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ca0ba0109087e891779' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ca2ba0109087a891603' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075caae80d3708c5a574bd' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cb286541a08fc14b8bf' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cb586541a08fc14b8c7' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cb7ba0109087e8917c2' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cb9ba0109087a891642' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102155f317620028737d52a' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-007/eeg/sub-007_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021564c7a976029b9e5c51' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cc986541a090014bcab' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102156ec7a976029b9e5c86' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021568317620028737d553' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ccee80d3708c5a574e1' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cc6ba0109087e8917f2' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cbce80d3708caa583ce' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ccbba0109087a89166f' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ce0ba0109087a89168a' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021577317620027c3848d6' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cd0e80d3708c5a574e9' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cd8ba0109087a891682' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cdbe80d3708caa5842b' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cdee80d3708caa58436' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-009/eeg/sub-009_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//61021572c7a976029b9e5ca8' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_electrodes.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//6102157d317620028737d5b5' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_events.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cefba0109087e89184b' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.set' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075cede80d3708c7a5817f' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_coordsystem.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//610215800c4cba0269bc9aa4' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.fdt' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ce986541a090014bd1b' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_channels.tsv' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ce1e80d3708caa5844e' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
Downloading file 'erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.json' from 'https://files.de-1.osf.io/v1/resources/pfde9/providers/osfstorage//60075ceb86541a08f914ab9e' to '/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data'.
files_dict
{'raw_files': ['/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.set'],
'log_files': ['/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_events.tsv']}
Then we’ll run a for loop with all the processing steps inside it.
Before the loop, we create empty lists where we will store the processed outputs (in this case, the evokeds) for all participants.
evokeds_face = []
evokeds_car = []
evokeds_diff = []
for raw_file, log_file in zip(files_dict['raw_files'],
files_dict['log_files']):
# Preprocessing
raw = read_raw(raw_file, preload=True)
raw = set_bipolar_reference(raw, anode='FP1', cathode='VEOG_lower',
ch_name='VEOG', drop_refs=False)
raw = set_bipolar_reference(raw, anode='HEOG_right', cathode='HEOG_left',
ch_name='HEOG', drop_refs=False)
raw = raw.set_channel_types({'VEOG': 'eog', 'HEOG': 'eog'})
raw = raw.drop_channels(['VEOG_lower', 'HEOG_right', 'HEOG_left'])
raw = raw.set_montage('biosemi64', match_case=False)
raw = raw.filter(l_freq=0.1, h_freq=30.0)
raw_copy = raw.copy().filter(l_freq=1.0, h_freq=None, verbose=False)
ica = ICA(n_components=15)
ica = ica.fit(raw_copy)
eog_indices, eog_scores = ica.find_bads_eog(raw, ch_name=['VEOG', 'HEOG'],
verbose=False)
ica.exclude = eog_indices
raw = ica.apply(raw)
raw = raw.set_eeg_reference('average')
# Epoching
log = pd.read_csv(log_file, sep='\t')
events = log[['sample', 'duration', 'value']].values.astype(int)
events = merge_events(events, ids=range(1, 41), new_id=1)
events = merge_events(events, ids=range(41, 81), new_id=2)
event_id = {'face': 1, 'car': 2}
epochs = Epochs(raw, events, event_id, tmin=-0.2, tmax=0.8,
baseline=(-0.2, 0.0), preload=True)
epochs = epochs.drop_bad({'eeg': 200e-6})
# Averaging
evoked_face = epochs['face'].average()
evokeds_face.append(evoked_face)
evoked_car = epochs['car'].average()
evokeds_car.append(evoked_car)
evoked_list = [evoked_face, evoked_car]
evoked_diff = combine_evoked(evoked_list, weights=[1, -1])
evokeds_diff.append(evoked_diff)
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.fdt
Reading 0 ... 699391 = 0.000 ... 682.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=699392
Range : 0 ... 699391 = 0.000 ... 682.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=699392
Range : 0 ... 699391 = 0.000 ... 682.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 7.1s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
Rejecting epoch based on EEG : ['P8', 'PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'PO7', 'P8', 'PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'P7', 'PO7', 'Oz', 'CPz', 'P8', 'PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
9 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.fdt
Reading 0 ... 683007 = 0.000 ... 666.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=683008
Range : 0 ... 683007 = 0.000 ... 666.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=683008
Range : 0 ... 683007 = 0.000 ... 666.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 10.1s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
0 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.fdt
Reading 0 ... 579583 = 0.000 ... 565.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=579584
Range : 0 ... 579583 = 0.000 ... 565.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=579584
Range : 0 ... 579583 = 0.000 ... 565.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 6.0s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
6 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.fdt
Reading 0 ... 649215 = 0.000 ... 633.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=649216
Range : 0 ... 649215 = 0.000 ... 633.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=649216
Range : 0 ... 649215 = 0.000 ... 633.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 5.1s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
Rejecting epoch based on EEG : ['O2']
1 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.fdt
Reading 0 ... 603135 = 0.000 ... 588.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=603136
Range : 0 ... 603135 = 0.000 ... 588.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=603136
Range : 0 ... 603135 = 0.000 ... 588.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 9.8s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
0 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.fdt
Reading 0 ... 529407 = 0.000 ... 516.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=529408
Range : 0 ... 529407 = 0.000 ... 516.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=529408
Range : 0 ... 529407 = 0.000 ... 516.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 6.3s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
Rejecting epoch based on EEG : ['FP2']
Rejecting epoch based on EEG : ['P10']
Rejecting epoch based on EEG : ['FP2']
3 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.fdt
Reading 0 ... 589823 = 0.000 ... 575.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=589824
Range : 0 ... 589823 = 0.000 ... 575.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=589824
Range : 0 ... 589823 = 0.000 ... 575.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 6.1s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
0 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.fdt
Reading 0 ... 804863 = 0.000 ... 785.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=804864
Range : 0 ... 804863 = 0.000 ... 785.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=804864
Range : 0 ... 804863 = 0.000 ... 785.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 11.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
0 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.fdt
Reading 0 ... 559103 = 0.000 ... 545.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=559104
Range : 0 ... 559103 = 0.000 ... 545.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=559104
Range : 0 ... 559103 = 0.000 ... 545.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 3.7s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
0 bad epochs dropped
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.fdt
Reading 0 ... 556031 = 0.000 ... 542.999 secs...
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=556032
Range : 0 ... 556031 = 0.000 ... 542.999 secs
Ready.
Added the following bipolar channels:
VEOG
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=1, n_times=556032
Range : 0 ... 556031 = 0.000 ... 542.999 secs
Ready.
Added the following bipolar channels:
HEOG
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 33793 samples (33.001 s)
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 7.7s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 1025 original time points ...
0 bad epochs dropped
0 bad epochs dropped
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.4s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.4s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.3s
We have collected the averaged ERP (evoked) for both conditions (face and car) from all participants. Averaging one more time, this time not across trials but across participants, gives us the grand average. Let’s do this separately for both condition and display the grand averages as a time course plot:
grand_evoked_face = combine_evoked(evokeds_face, weights='equal')
grand_evoked_face.comment = 'face'
grand_evoked_car = combine_evoked(evokeds_car, weights='equal')
grand_evoked_car.comment = 'car'
grand_evoked_list = [grand_evoked_face, grand_evoked_car]
_ = plot_compare_evokeds(grand_evoked_list, picks='PO8')
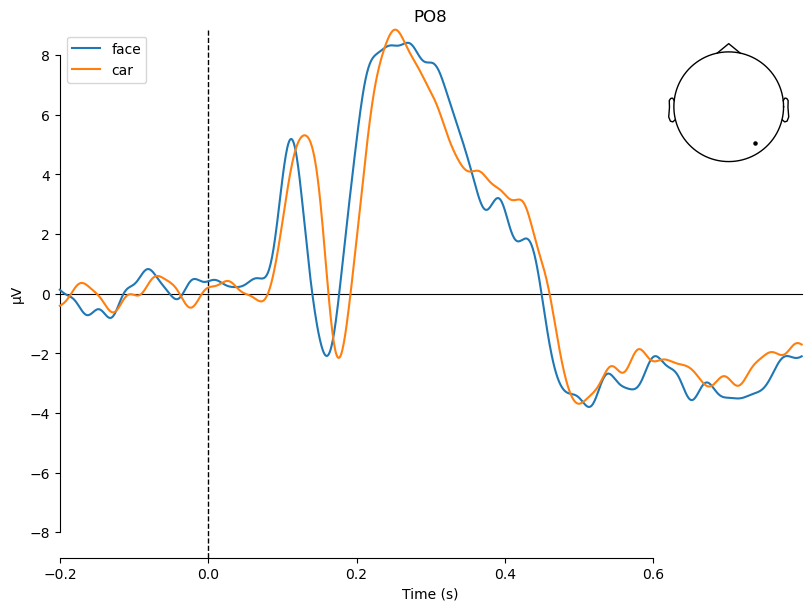
We see that the first negative peak in the ERP (N1/N170 component) is earlier and (and maybe also larger) for faces than for cars.
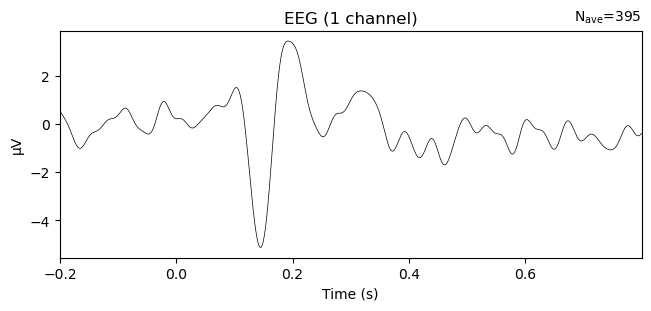
This becomes even more apparent when grand-averaging and plotting the difference waves:
grand_evoked_diff = combine_evoked(evokeds_diff, weights='equal')
grand_evoked_diff.comment = 'face - car'
_ = grand_evoked_diff.plot(picks='PO8')
Need more than one channel to make topography for eeg. Disabling interactivity.
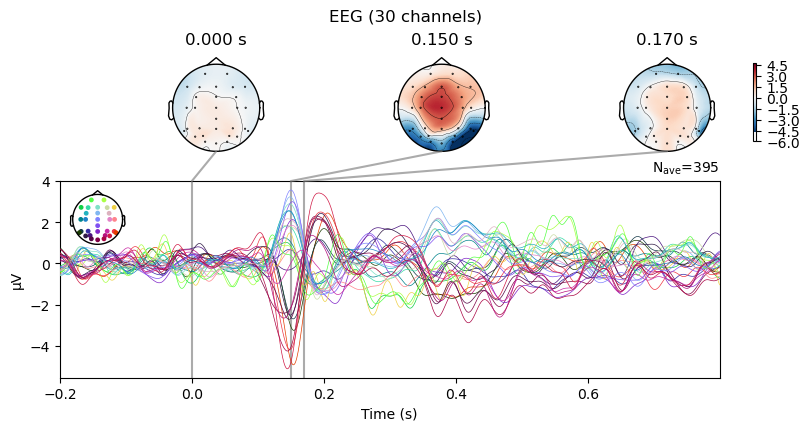
The corresponding butterfly and scalp topography plot looks like this:
_ = grand_evoked_diff.plot_joint(times=[0.0, 0.15, 0.17])
No projector specified for this dataset. Please consider the method self.add_proj.
5.3. Neuro Lab pipeline#
At the Abdel Rahman Lab for Neurocognitive Psychology at HU Berlin (the “Neuro Lab,” for short), we’ve developed a Python package that provides a fully automated EEG processing pipeline. The pipeline was originally developed and published in the MATLAB language by Frömer et al (2018). The more recent Python version is available at https://hu-neuro-pipeline.readthedocs.io.
The pipeline is fully automated in the sense that it takes raw EEG data from multiple participants as an input and performs a number of standardized processing step at the participant and group level with minimal user input. The typical steps are visualized in this flowchart:
Fig. 5.1 Processing steps in the hu-neuro-pipeline package.
Source: Docs.#
The pipeline package only has one main function, called group_pipeline().
Let’s use it to process the same example data as before:
trials, evokeds, config = group_pipeline(raw_files=files_dict['raw_files'],
log_files=files_dict['log_files'],
output_dir='output',
montage='biosemi64',
ica_method='fastica',
ica_n_components=15,
triggers=range(1, 81),
skip_log_conditions={'value': range(81, 203)},
components={'name': 'N170',
'tmin': 0.11,
'tmax': 0.15,
'roi': ['PO8']},
average_by={'face': 'value <= 40',
'car': 'value > 40'})
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.fdt
Reading 0 ... 699391 = 0.000 ... 682.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 7.3s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['Oz', 'F8', 'P8', 'PO8']
Rejecting epoch based on EEG : ['PO7', 'P8', 'PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['FP2', 'F4', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F7', 'PO8']
Rejecting epoch based on EEG : ['PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'P7', 'PO7', 'F8', 'P8', 'PO8']
Rejecting epoch based on EEG : ['PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'P7', 'PO7', 'Oz', 'CPz', 'F8', 'P8', 'PO8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F4', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
87 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.fdt
Reading 0 ... 683007 = 0.000 ... 666.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 14.1s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['P3']
1 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.fdt
Reading 0 ... 579583 = 0.000 ... 565.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 14.4s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
3 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.fdt
Reading 0 ... 649215 = 0.000 ... 633.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 9.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['PO7', 'PO8', 'O2']
Rejecting epoch based on EEG : ['O2']
Rejecting epoch based on EEG : ['F3']
4 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.fdt
Reading 0 ... 603135 = 0.000 ... 588.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 10.2s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.fdt
Reading 0 ... 529407 = 0.000 ... 516.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 5.6s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['F7', 'FP2']
Rejecting epoch based on EEG : ['P10']
Rejecting epoch based on EEG : ['F4']
Rejecting epoch based on EEG : ['FP2']
4 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.fdt
Reading 0 ... 589823 = 0.000 ... 575.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 10.2s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.fdt
Reading 0 ... 804863 = 0.000 ... 785.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 17.6s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.fdt
Reading 0 ... 559103 = 0.000 ... 545.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 5.8s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.fdt
Reading 0 ... 556031 = 0.000 ... 542.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 8.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['C5']
Rejecting epoch based on EEG : ['F7', 'F8']
Rejecting epoch based on EEG : ['F7']
Rejecting epoch based on EEG : ['F8']
4 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Processing group level ===
Identifying common channels ...
Identifying common channels ...
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
In this example we’ve specified:
The input and output file paths (
raw_files,log_files,output_dir)Some preprocessing options (
montage,ica_method,ica_n_components; note that the pipeline also applies a 0.1–40 Hz band-pass filter by default)The event codes of interest for epoching (
triggers; note that the pipeline also applies a default peak-to-peak rejection at 200 µV)Some event codes to skip (
skip_log_conditions; anything other than faces and houses)The definition of our ERP component(s) of interest (
components; here only the N170)Rules to create by-participant condition averages/evokeds (
average_by; here for faces and cars)
A (long) list of these and all other input options is available on the pipeline documentation website.
The pipeline returns three objects (which also get written as text files into the output_dir):
trials: A data frame with the single trial data for all participants, also containing the single trial ERP amplitudes (averaged across the time window and channels of interest)evokeds: A data frame with the averaged (evoked) ERP amplitudes for all time points, channels, and participantsconfig: A dictionary with the pipeline configuration
Let’s look of each of these in turn:
5.3.1. Single trial data#
trials
| participant_id | onset | duration | sample | trial_type | stim_file | value | N170 | |
|---|---|---|---|---|---|---|---|---|
| 0 | sub-001_task-N170_eeg | 15.0498 | 0.3 | 15412 | stimulus | NaN | 79 | 0.298910 |
| 1 | sub-001_task-N170_eeg | 16.5986 | 0.3 | 16998 | stimulus | NaN | 73 | 12.099460 |
| 2 | sub-001_task-N170_eeg | 18.0322 | 0.3 | 18466 | stimulus | NaN | 75 | 1.478766 |
| 3 | sub-001_task-N170_eeg | 21.1641 | 0.3 | 21673 | stimulus | NaN | 17 | 6.740304 |
| 4 | sub-001_task-N170_eeg | 22.7305 | 0.3 | 23277 | stimulus | NaN | 32 | -1.743442 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1595 | sub-010_task-N170_eeg | 523.1045 | 0.3 | 535660 | stimulus | NaN | 17 | 12.973517 |
| 1596 | sub-010_task-N170_eeg | 526.1035 | 0.3 | 538731 | stimulus | NaN | 71 | 14.055316 |
| 1597 | sub-010_task-N170_eeg | 529.0850 | 0.3 | 541784 | stimulus | NaN | 4 | -14.064324 |
| 1598 | sub-010_task-N170_eeg | 530.6348 | 0.3 | 543371 | stimulus | NaN | 22 | -1.718830 |
| 1599 | sub-010_task-N170_eeg | 533.6338 | 0.3 | 546442 | stimulus | NaN | 43 | 4.418730 |
1600 rows × 8 columns
We could (and will!) use this for statistically analyzing the ERP component(s) of interest. Specifically, we can fit a mixed-effects model that tests if the single trial N170 amplitudes differs as a function of the condition of the trial (face vs. car).
5.3.2. Evokeds#
evokeds
| participant_id | label | query | time | FP1 | F3 | F7 | FC3 | C3 | C5 | ... | Cz | C4 | C6 | P4 | P8 | P10 | PO8 | PO4 | O2 | N170 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | sub-001_task-N170_eeg | face | value <= 40 | -0.500000 | 5.789564 | -0.034750 | 1.660734 | -1.520637 | -1.074939 | -0.738493 | ... | -1.048279 | -1.561112 | -1.006365 | -1.609887 | 1.768214 | 2.574583 | 0.807337 | -1.772100 | 1.039566 | 0.807337 |
| 1 | sub-001_task-N170_eeg | face | value <= 40 | -0.499023 | 5.680271 | -0.100856 | 1.595741 | -1.594228 | -1.011824 | -0.663110 | ... | -1.086347 | -1.568407 | -1.134829 | -1.396554 | 1.590214 | 2.462847 | 0.594162 | -1.514759 | 0.947991 | 0.594162 |
| 2 | sub-001_task-N170_eeg | face | value <= 40 | -0.498047 | 5.492423 | -0.201391 | 1.515443 | -1.676766 | -0.951158 | -0.600500 | ... | -1.124984 | -1.579886 | -1.221997 | -1.183525 | 1.420789 | 2.362887 | 0.417310 | -1.246389 | 0.868075 | 0.417310 |
| 3 | sub-001_task-N170_eeg | face | value <= 40 | -0.497070 | 5.234354 | -0.334659 | 1.427033 | -1.766560 | -0.895557 | -0.553428 | ... | -1.163056 | -1.595503 | -1.272581 | -0.980823 | 1.272416 | 2.279901 | 0.288978 | -0.977250 | 0.807886 | 0.288978 |
| 4 | sub-001_task-N170_eeg | face | value <= 40 | -0.496094 | 4.916389 | -0.496533 | 1.336095 | -1.860806 | -0.847166 | -0.524221 | ... | -1.199010 | -1.614431 | -1.293715 | -0.797415 | 1.155798 | 2.217527 | 0.218337 | -0.717113 | 0.774866 | 0.218337 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 40955 | sub-010_task-N170_eeg | car | value > 40 | 1.495117 | 1.696184 | -1.211245 | 0.285654 | -0.857636 | -1.820990 | -3.490791 | ... | -0.625288 | -0.666407 | 0.613045 | -0.259044 | 2.407768 | 3.256570 | 1.626962 | -0.318652 | 1.387889 | 1.626962 |
| 40956 | sub-010_task-N170_eeg | car | value > 40 | 1.496094 | 1.662606 | -1.163206 | 0.330227 | -0.782776 | -1.747530 | -3.317312 | ... | -0.579745 | -0.660078 | 0.606120 | -0.278228 | 2.331428 | 3.123536 | 1.553047 | -0.378411 | 1.313106 | 1.553047 |
| 40957 | sub-010_task-N170_eeg | car | value > 40 | 1.497070 | 1.627197 | -1.113622 | 0.386814 | -0.702416 | -1.660422 | -3.107137 | ... | -0.534325 | -0.659212 | 0.590221 | -0.302836 | 2.249132 | 2.984847 | 1.472701 | -0.436253 | 1.230004 | 1.472701 |
| 40958 | sub-010_task-N170_eeg | car | value > 40 | 1.498047 | 1.591662 | -1.063696 | 0.452267 | -0.617246 | -1.561477 | -2.868092 | ... | -0.489355 | -0.663735 | 0.564956 | -0.332295 | 2.161821 | 2.844188 | 1.387976 | -0.491059 | 1.140647 | 1.387976 |
| 40959 | sub-010_task-N170_eeg | car | value > 40 | 1.499023 | 1.557830 | -1.014824 | 0.523435 | -0.528657 | -1.453341 | -2.609703 | ... | -0.445078 | -0.673118 | 0.530453 | -0.365673 | 2.070765 | 2.705437 | 1.301141 | -0.541826 | 1.047361 | 1.301141 |
40960 rows × 35 columns
The evokeds can be used for plotting the ERP time courses for both conditions. For this, we will use the seaborn package, which is great for visualizing tabular data (similar to the ggplot2 package in R).
_ = sns.lineplot(evokeds, x='time', y='PO8', hue='label',
estimator='mean', errorbar='se')
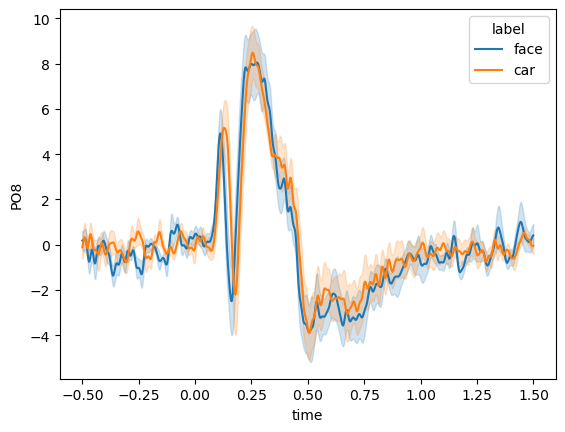
We can make the plot yet a bit prettier by adding custom x-axis limits, vertical and horizontal lines at zero, and more informative axis labels:
_ = sns.lineplot(evokeds, x='time', y='PO8', hue='label',
estimator='mean', errorbar='se')
_ = plt.margins(x=0.0, y=0.1)
_ = plt.xlim(-0.2, 0.8)
_ = plt.axvline(0.0, color='black', linestyle='--')
_ = plt.axhline(0.0, color='black', linestyle='--')
_ = plt.xlabel('Time (s)')
_ = plt.ylabel('PO8 amplitude (µV)')
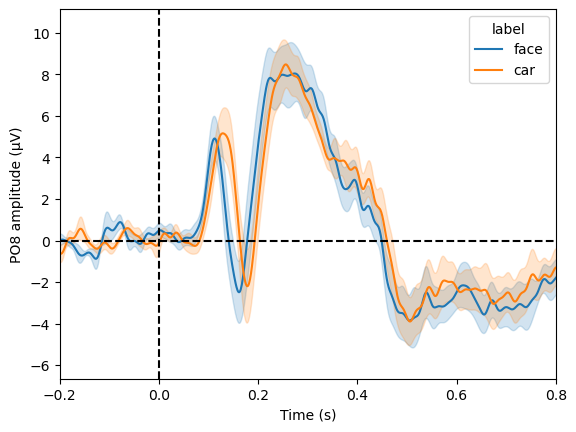
5.3.3. Pipeline configuration#
Finally, the config output is a dictionary with information about the pipeline run.
It contains all the user-specified and default input arguments plus some new information that the pipeline has computed along the way (e.g., 'auto_rejected_epochs', the rejected epochs for each participant):
config
{'raw_files': ['/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.set',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.set'],
'log_files': ['/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_events.tsv',
'/home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_events.tsv'],
'output_dir': 'output',
'clean_dir': None,
'epochs_dir': None,
'report_dir': None,
'to_df': True,
'downsample_sfreq': None,
'veog_channels': 'auto',
'heog_channels': 'auto',
'montage': 'biosemi64',
'bad_channels': [None, None, None, None, None, None, None, None, None, None],
'ref_channels': 'average',
'besa_files': [None, None, None, None, None, None, None, None, None, None],
'ica_method': 'fastica',
'ica_n_components': 15,
'highpass_freq': 0.1,
'lowpass_freq': 40.0,
'triggers': 'range(1, 81)',
'triggers_column': None,
'epochs_tmin': -0.5,
'epochs_tmax': 1.5,
'baseline': (-0.2, 0.0),
'skip_log_rows': [None, None, None, None, None, None, None, None, None, None],
'skip_log_conditions': {'value': 'range(81, 203)'},
'reject_peak_to_peak': 200.0,
'components': {'name': ['N170'],
'tmin': [0.11],
'tmax': [0.15],
'roi': ['PO8']},
'average_by': {'face': 'value <= 40', 'car': 'value > 40'},
'perform_tfr': False,
'tfr_subtract_evoked': False,
'tfr_freqs': [4.0,
5.0,
6.0,
7.0,
8.0,
9.0,
10.0,
11.0,
12.0,
13.0,
14.0,
15.0,
16.0,
17.0,
18.0,
19.0,
20.0,
21.0,
22.0,
23.0,
24.0,
25.0,
26.0,
27.0,
28.0,
29.0,
30.0,
31.0,
32.0,
33.0,
34.0,
35.0,
36.0,
37.0,
38.0,
39.0,
40.0],
'tfr_cycles': [2.0,
2.5,
3.0,
3.5,
4.0,
4.5,
5.0,
5.5,
6.0,
6.5,
7.0,
7.5,
8.0,
8.5,
9.0,
9.5,
10.0,
10.5,
11.0,
11.5,
12.0,
12.5,
13.0,
13.5,
14.0,
14.5,
15.0,
15.5,
16.0,
16.5,
17.0,
17.5,
18.0,
18.5,
19.0,
19.5,
20.0],
'tfr_mode': 'percent',
'tfr_baseline': (-0.45, -0.05),
'tfr_components': {'name': [],
'tmin': [],
'tmax': [],
'fmin': [],
'fmax': [],
'roi': []},
'perm_contrasts': [],
'perm_tmin': 0.0,
'perm_tmax': 1.0,
'perm_channels': None,
'perm_fmin': None,
'perm_fmax': None,
'n_jobs': 1,
'vhdr_files': None,
'auto_rejected_epochs': {'sub-001_task-N170_eeg': [32,
33,
34,
35,
36,
41,
42,
43,
46,
47,
48,
49,
50,
52,
53,
54,
55,
56,
61,
62,
63,
66,
67,
68,
69,
70,
71,
72,
73,
74,
82,
83,
84,
85,
92,
93,
94,
95,
96,
97,
98,
99,
100,
101,
105,
107,
108,
109,
110,
112,
113,
114,
115,
116,
117,
118,
119,
120,
121,
122,
124,
125,
126,
127,
128,
129,
131,
132,
133,
134,
135,
136,
142,
143,
144,
145,
146,
147,
148,
149,
150,
153,
154,
155,
156,
157,
158],
'sub-002_task-N170_eeg': [56],
'sub-003_task-N170_eeg': [42, 66, 87],
'sub-004_task-N170_eeg': [35, 96, 97, 98],
'sub-005_task-N170_eeg': [],
'sub-006_task-N170_eeg': [0, 45, 107, 131],
'sub-007_task-N170_eeg': [],
'sub-008_task-N170_eeg': [],
'sub-009_task-N170_eeg': [],
'sub-010_task-N170_eeg': [15, 49, 50, 85]},
'auto_ica_bad_components': {'sub-001_task-N170_eeg': [0],
'sub-002_task-N170_eeg': [0, 6],
'sub-003_task-N170_eeg': [0, 7],
'sub-004_task-N170_eeg': [0],
'sub-005_task-N170_eeg': [0, 2],
'sub-006_task-N170_eeg': [0],
'sub-007_task-N170_eeg': [0, 1],
'sub-008_task-N170_eeg': [0, 12],
'sub-009_task-N170_eeg': [0, 2],
'sub-010_task-N170_eeg': [0, 1]},
'package_versions': {'python': '3.11.7',
'pipeline': '0.8.3',
'mne': '1.6.0',
'numpy': '1.26.2',
'pandas': '2.1.4',
'scikit-learn': '1.3.2'}}
5.4. Exercises#
Run the EEG analysis pipeline (using a custom
forloop or thegroup_pipeline()function) for 10 participants from a different ERP CORE experiment (valid experiment names are'N170','MMN','N2pc','N400','P3', or'ERN'). Create evokeds for the two conditions of interest and visualize their time course.
# Your code goes here
...
5.5. Further reading#
Paper Group-level EEG-processing pipeline for flexible single trial-based analyses including linear mixed models (Frömer et al, 2018)
hu-neuro-pipeline package documentation
Slides on the hu-neuro-pipeline package
5.6. References#
- Fromer et al., 2018(1,2)
Frömer, R., Maier, M., & Abdel Rahman, R. (2018). Group-level EEG-processing pipeline for flexible single trial-based analyses including linear mixed models. Frontiers in Neuroscience, 12, 48. doi:10.3389/fnins.2018.00048
- Gramfort et al., 2013
Gramfort, A., Luessi, M., Larson, E., Engemann, D. A., Strohmeier, D., Brodbeck, C., … Hämäläinen, M. (2013). MEG and EEG data analysis with MNE-Python. Frontiers in Neuroscience, 7, 267. doi:10.3389/fnins.2013.00267


