6. Statistics#
In the previous chapters, we already got an idea about the differences in EEG activity between our conditions of interest (e.g., faces versus cars), as shown in the time course and scalp topography plots. However, we have to model the data statistically to be able to quantify the size of this difference and test if it is statistically significant (i.e., not due to chance). In this chapter, we will encounter different statistical tests that can be used to do this.
While the EEG processing is done in Python as before, we will use the R programming language for statistical modeling because it has a larger number of statistical functions and packages, and because it is widely used in the psychological science community.
Goals
Test for condition differences using “classical” models based on averaged data (e.g., \(t\)-tests, ANOVA)
Do the same by applying linear mixed models to the single trial data
6.1. Load Python packages#
We’ll use the hu-neuro-pipeline package (introduced in Chapter 5) for EEG processing, and Numpy and seaborn for post-processing and plotting. As mentioned before, the actual statistical modeling will be done in R, but there are also Python packages for this (e.g., statsmodels).
# %pip install numpy seaborn hu-neuro-pipeline
import numpy as np
import seaborn as sns
from pipeline import group_pipeline
from pipeline.datasets import get_erpcore
6.2. Re-run the pipeline#
We use the same processing pipeline as introduced in the Neuro Lab pipeline section of the previous chapter, giving us the single trial data and the average time courses.
files_dict = get_erpcore('N170', participants=10, path='data')
## %% tags=["output_scroll"]
trials, evokeds, config = group_pipeline(raw_files=files_dict['raw_files'],
log_files=files_dict['log_files'],
output_dir='output',
montage='biosemi64',
ica_method='fastica',
ica_n_components=15,
triggers=range(1, 81),
skip_log_conditions={'value': range(81, 203)},
components={'name': 'N170',
'tmin': 0.110,
'tmax': 0.150,
'roi': ['PO8']},
average_by={'face': 'value <= 40',
'car': 'value > 40'})
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-001/eeg/sub-001_task-N170_eeg.fdt
Reading 0 ... 699391 = 0.000 ... 682.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 7.4s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['Oz', 'F8', 'P8', 'PO8']
Rejecting epoch based on EEG : ['PO7', 'P8', 'PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['FP2', 'F4', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F7', 'PO8']
Rejecting epoch based on EEG : ['PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'P7', 'PO7', 'F8', 'P8', 'PO8']
Rejecting epoch based on EEG : ['PO8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['C5', 'P7', 'PO7', 'Oz', 'CPz', 'F8', 'P8', 'PO8']
Rejecting epoch based on EEG : ['C5', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F4', 'F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['F8']
87 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-002/eeg/sub-002_task-N170_eeg.fdt
Reading 0 ... 683007 = 0.000 ... 666.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 14.1s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['P3']
1 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-003/eeg/sub-003_task-N170_eeg.fdt
Reading 0 ... 579583 = 0.000 ... 565.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 14.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
Rejecting epoch based on EEG : ['FP1']
3 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-004/eeg/sub-004_task-N170_eeg.fdt
Reading 0 ... 649215 = 0.000 ... 633.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 9.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['F8']
Rejecting epoch based on EEG : ['PO7', 'PO8', 'O2']
Rejecting epoch based on EEG : ['O2']
Rejecting epoch based on EEG : ['F3']
4 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-005/eeg/sub-005_task-N170_eeg.fdt
Reading 0 ... 603135 = 0.000 ... 588.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 10.2s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-006/eeg/sub-006_task-N170_eeg.fdt
Reading 0 ... 529407 = 0.000 ... 516.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 5.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 1 ICA component
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['F7', 'FP2']
Rejecting epoch based on EEG : ['P10']
Rejecting epoch based on EEG : ['F4']
Rejecting epoch based on EEG : ['FP2']
4 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-007/eeg/sub-007_task-N170_eeg.fdt
Reading 0 ... 589823 = 0.000 ... 575.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 10.2s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-008/eeg/sub-008_task-N170_eeg.fdt
Reading 0 ... 804863 = 0.000 ... 785.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 17.5s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-009/eeg/sub-009_task-N170_eeg.fdt
Reading 0 ... 559103 = 0.000 ... 545.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 5.8s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
0 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Reading raw data from /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.set ===
Reading /home/runner/work/intro-to-eeg/intro-to-eeg/ipynb/data/erpcore/N170/sub-010/eeg/sub-010_task-N170_eeg.fdt
Reading 0 ... 556031 = 0.000 ... 542.999 secs...
Adding bipolar channel VEOG (FP1 - VEOG_lower)
Adding bipolar channel HEOG (HEOG_left - HEOG_right)
Loading standard montage biosemi64
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom ('EEG',) reference.
Fitting ICA to data using 30 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 8.6s.
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 30 PCA components
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 0.1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.10
- Lower transition bandwidth: 0.10 Hz (-6 dB cutoff frequency: 0.05 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 33793 samples (33.001 s)
Not setting metadata
160 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 160 events and 2049 original time points ...
0 bad epochs dropped
<Epochs | 160 events (all good), -0.5 - 1.49902 s, baseline -0.2 - 0 s, ~87.5 MB, data loaded,
'1': 2
'10': 2
'11': 2
'12': 2
'13': 2
'14': 2
'15': 2
'16': 2
'17': 2
'18': 2
and 70 more events ...>
Adding metadata with 6 columns
Rejecting epoch based on EEG : ['C5']
Rejecting epoch based on EEG : ['F7', 'F8']
Rejecting epoch based on EEG : ['F7']
Rejecting epoch based on EEG : ['F8']
4 bad epochs dropped
Computing single trial ERP amplitudes for 'N170'
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
=== Processing group level ===
Identifying common channels ...
Identifying common channels ...
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:103: UserWarning: Converting `ica_n_components` to integer: 15 -> 15
warn(f'Converting `ica_n_components` to integer: {n_components} -> ' +
/home/runner/micromamba/envs/intro-to-eeg/lib/python3.11/site-packages/pipeline/preprocessing.py:115: FutureWarning: The default for pick_channels will change from ordered=False to ordered=True in 1.5 and this will result in a change of behavior because the resulting channel order will not match. Either use a channel order that matches your instance or pass ordered=False.
eog_indices, _ = ica.find_bads_eog(
6.3. Single trial data#
The main output of the hu-neuro-pipeline package is the single trial data frame, which contains the EEG data for each trial (column N170), averaged across an a priori hypothesized time window and electrode(s) of interest (see the components argument above).
trials
| participant_id | onset | duration | sample | trial_type | stim_file | value | N170 | |
|---|---|---|---|---|---|---|---|---|
| 0 | sub-001_task-N170_eeg | 15.0498 | 0.3 | 15412 | stimulus | NaN | 79 | 0.298910 |
| 1 | sub-001_task-N170_eeg | 16.5986 | 0.3 | 16998 | stimulus | NaN | 73 | 12.099460 |
| 2 | sub-001_task-N170_eeg | 18.0322 | 0.3 | 18466 | stimulus | NaN | 75 | 1.478766 |
| 3 | sub-001_task-N170_eeg | 21.1641 | 0.3 | 21673 | stimulus | NaN | 17 | 6.740304 |
| 4 | sub-001_task-N170_eeg | 22.7305 | 0.3 | 23277 | stimulus | NaN | 32 | -1.743442 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1595 | sub-010_task-N170_eeg | 523.1045 | 0.3 | 535660 | stimulus | NaN | 17 | 12.973517 |
| 1596 | sub-010_task-N170_eeg | 526.1035 | 0.3 | 538731 | stimulus | NaN | 71 | 14.055316 |
| 1597 | sub-010_task-N170_eeg | 529.0850 | 0.3 | 541784 | stimulus | NaN | 4 | -14.064324 |
| 1598 | sub-010_task-N170_eeg | 530.6348 | 0.3 | 543371 | stimulus | NaN | 22 | -1.718830 |
| 1599 | sub-010_task-N170_eeg | 533.6338 | 0.3 | 546442 | stimulus | NaN | 43 | 4.418730 |
1600 rows × 8 columns
Using a combination of pandas and Numpy, we’ll create a new column in the data frame with verbal labels for our two conditions of interest (faces and cars).
This is based on the numerical event codes (stored in the value column), the meaning of which was described in the Extract events section of the Epoching chapter.
trials['condition'] = np.where(trials['value'] <= 40, 'face', 'car')
trials
| participant_id | onset | duration | sample | trial_type | stim_file | value | N170 | condition | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | sub-001_task-N170_eeg | 15.0498 | 0.3 | 15412 | stimulus | NaN | 79 | 0.298910 | car |
| 1 | sub-001_task-N170_eeg | 16.5986 | 0.3 | 16998 | stimulus | NaN | 73 | 12.099460 | car |
| 2 | sub-001_task-N170_eeg | 18.0322 | 0.3 | 18466 | stimulus | NaN | 75 | 1.478766 | car |
| 3 | sub-001_task-N170_eeg | 21.1641 | 0.3 | 21673 | stimulus | NaN | 17 | 6.740304 | face |
| 4 | sub-001_task-N170_eeg | 22.7305 | 0.3 | 23277 | stimulus | NaN | 32 | -1.743442 | face |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1595 | sub-010_task-N170_eeg | 523.1045 | 0.3 | 535660 | stimulus | NaN | 17 | 12.973517 | face |
| 1596 | sub-010_task-N170_eeg | 526.1035 | 0.3 | 538731 | stimulus | NaN | 71 | 14.055316 | car |
| 1597 | sub-010_task-N170_eeg | 529.0850 | 0.3 | 541784 | stimulus | NaN | 4 | -14.064324 | face |
| 1598 | sub-010_task-N170_eeg | 530.6348 | 0.3 | 543371 | stimulus | NaN | 22 | -1.718830 | face |
| 1599 | sub-010_task-N170_eeg | 533.6338 | 0.3 | 546442 | stimulus | NaN | 43 | 4.418730 | car |
1600 rows × 9 columns
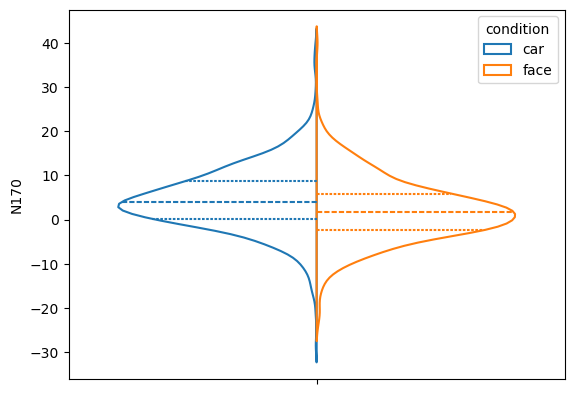
Using seaborn, we can plot the distribution of the single trial N170 amplitudes, separately for the two conditions. Note that this plot does not take into account the repeated measurements of the same participant, which we will address later.
_ = sns.violinplot(data=trials, y='N170', hue='condition',
inner='quart', split=True, fill=False)
6.4. Linear models#
The “traditional” way for statistical analysis of ERPs is to (a) average the data across trials for each participant and condition, and (b) apply a statistical test to the averaged data.
Let’s start with the first step.
The pandas package has the necessary methods to group the data by participant and condition (groupby() method), and compute the average N170 amplitude across trials for each grouping (mean() method).
trials_ave = trials[['participant_id', 'condition', 'N170']].\
groupby(['participant_id', 'condition']).\
mean().\
reset_index()
trials_ave
| participant_id | condition | N170 | |
|---|---|---|---|
| 0 | sub-001_task-N170_eeg | car | 4.717367 |
| 1 | sub-001_task-N170_eeg | face | 4.573434 |
| 2 | sub-002_task-N170_eeg | car | 7.233116 |
| 3 | sub-002_task-N170_eeg | face | 6.399796 |
| 4 | sub-003_task-N170_eeg | car | 12.412293 |
| 5 | sub-003_task-N170_eeg | face | 9.789007 |
| 6 | sub-004_task-N170_eeg | car | 1.016570 |
| 7 | sub-004_task-N170_eeg | face | -0.961611 |
| 8 | sub-005_task-N170_eeg | car | 4.486981 |
| 9 | sub-005_task-N170_eeg | face | 1.418827 |
| 10 | sub-006_task-N170_eeg | car | 2.207928 |
| 11 | sub-006_task-N170_eeg | face | -0.828506 |
| 12 | sub-007_task-N170_eeg | car | 3.189184 |
| 13 | sub-007_task-N170_eeg | face | -0.297194 |
| 14 | sub-008_task-N170_eeg | car | 4.791464 |
| 15 | sub-008_task-N170_eeg | face | 1.105288 |
| 16 | sub-009_task-N170_eeg | car | 2.737309 |
| 17 | sub-009_task-N170_eeg | face | 0.263003 |
| 18 | sub-010_task-N170_eeg | car | 3.380139 |
| 19 | sub-010_task-N170_eeg | face | -0.360485 |
Now we can pass the data frame to R and apply an appropriate statistical test.
Using the rpy2 package, we can run R code directly in the Jupyter notebook, using the %%R magic command at the beginning of a code cell.
%load_ext rpy2.ipython
An appropriate statistical test needs to take into account that our two conditions are manipulated within participants, that is, we have repeated measures that are likely correlated with one another (violating the independence assumption of many statistical tests, e.g., linear regression).
Luckily, there are statistical tests that can handle repeated measures data, such as the paired \(t\)-test or repeated measures ANOVA.
Let’s start with the paired \(t\)-test:
%%R -i trials_ave
t.test(N170 ~ condition, data = trials_ave, paired = TRUE)
Paired t-test
data: N170 by condition
t = 6.5598, df = 9, p-value = 0.000104
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
1.642506 3.371652
sample estimates:
mean difference
2.507079
We see that in this sample, the amplitude in response to faces is approximately 2.51 µV lower (more negative) than in response to cars, as would be expected for the N170 component. This difference is statistically significant with \(t(9) = -6.56\), \(p \approx .0001\).
Note that we could have gotten the same result by applying a one sample \(t\)-test to the difference scores:
trials_ave_wide = trials_ave.pivot(index='participant_id', columns='condition',
values='N170')
trials_ave_wide['diff'] = trials_ave_wide['car'] - trials_ave_wide['face']
trials_ave_wide
| condition | car | face | diff |
|---|---|---|---|
| participant_id | |||
| sub-001_task-N170_eeg | 4.717367 | 4.573434 | 0.143933 |
| sub-002_task-N170_eeg | 7.233116 | 6.399796 | 0.833320 |
| sub-003_task-N170_eeg | 12.412293 | 9.789007 | 2.623286 |
| sub-004_task-N170_eeg | 1.016570 | -0.961611 | 1.978181 |
| sub-005_task-N170_eeg | 4.486981 | 1.418827 | 3.068154 |
| sub-006_task-N170_eeg | 2.207928 | -0.828506 | 3.036434 |
| sub-007_task-N170_eeg | 3.189184 | -0.297194 | 3.486378 |
| sub-008_task-N170_eeg | 4.791464 | 1.105288 | 3.686177 |
| sub-009_task-N170_eeg | 2.737309 | 0.263003 | 2.474306 |
| sub-010_task-N170_eeg | 3.380139 | -0.360485 | 3.740624 |
%%R -i trials_ave_wide
t.test(trials_ave_wide$diff)
One Sample t-test
data: trials_ave_wide$diff
t = 6.5598, df = 9, p-value = 0.000104
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
1.642506 3.371652
sample estimates:
mean of x
2.507079
Or by running a repeated measures ANOVA with a single (two-level, within-participant) factor:
%%R -i trials_ave
# install.packages("ez")
ez::ezANOVA(
data = trials_ave,
dv = N170,
wid = participant_id,
within = condition
)
$ANOVA
Effect DFn DFd F p p<.05 ges
2 condition 1 9 43.03072 0.0001039885 * 0.1291878
R[write to console]: Warning:
R[write to console]: Converting "participant_id" to factor for ANOVA.
R[write to console]: Warning:
R[write to console]: Converting "condition" to factor for ANOVA.
6.5. Linear mixed models#
The above approach of running a repeated measures linear model (ANOVA or \(t\)-test) on the averaged data is the “traditional” way of analyzing ERPs and still widely used. However, it comes with a number of drawbacks:
The averaging step discards a lot of information
We often don’t just have repeated measures of the same participants, but also of the same items (again violating the independence assumption) (Bürki et al, 2018, Judd et al, 2012)
We cannot include any information about specific trials or stimuli in the model
We cannot include any continuous predictor variables in the model
These models assume the same noise level (i.e., number of averaged trials) for all participants and conditions
A more flexible approach that can solve all of these problems (and more!) is to use a linear-mixed effects model (LMM) (Frömer et al, 2018, Volpert-Esmond et al, 2021). This model predicts the single trial amplitudes directly and accounts for repeated measures of participants and/or items by including random effects for these factors. They can also include continuous predictor variables at the participant and trial level, and they do not required a balanced design (i.e., the same number of trials for each participant and condition).
In R, we can use the lmer() function from the lme4 package to fit LMMs (Bates et al, 2015):
%%R -i trials
# install.packages("lme4")
mod <- lme4::lmer(N170 ~ 1 + condition + (1 | participant_id), trials)
summary(mod)
Linear mixed model fit by REML ['lmerMod']
Formula: N170 ~ 1 + condition + (1 | participant_id)
Data: trials
REML criterion at convergence: 9845.9
Scaled residuals:
Min 1Q Median 3Q Max
-4.8893 -0.5595 0.0214 0.5701 6.2065
Random effects:
Groups Name Variance Std.Dev.
participant_id (Intercept) 11.16 3.340
Residual 41.13 6.414
Number of obs: 1497, groups: participant_id, 10
Fixed effects:
Estimate Std. Error t value
(Intercept) 4.6762 1.0825 4.320
conditionface -2.6422 0.3316 -7.969
Correlation of Fixed Effects:
(Intr)
conditionfc -0.152
In this model, we’re predicting the single trial N170 amplitude (N170) from a categorical predictor variable (condition) and an intercept (1) as fixed effects.
We also specify a random intercept for the participant factor ((1 | participant_id)), which accounts for differences in the (average) voltage level between participants.
Note that we could (and should) also include a random slope for the condition factor ((1 + condition | participant_id)), as well as a random intercept for the item factor ((1 | value)).
However, in this example case with only 10 participants, this model would be overly complex and likely fail to converge.
In the above output, you will not any find any \(p\)-values to decide if the fixed effects are statistically significant.
If and how best to compute \(p\)-values for LMMs is still a matter of debate, but one common solution is a method called the Satterthwaite approximation.
This is implemented in the lmerTest package (Kuznetsova et al, 2017), which has a drop-in replacement for the lmer() function but with \(p\)-values:
%%R -i trials
# install.packages("lmerTest")
mod <- lmerTest::lmer(N170 ~ 1 + condition + (1 | participant_id), trials)
summary(mod)
Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: N170 ~ 1 + condition + (1 | participant_id)
Data: trials
REML criterion at convergence: 9845.9
Scaled residuals:
Min 1Q Median 3Q Max
-4.8893 -0.5595 0.0214 0.5701 6.2065
Random effects:
Groups Name Variance Std.Dev.
participant_id (Intercept) 11.16 3.340
Residual 41.13 6.414
Number of obs: 1497, groups: participant_id, 10
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 4.6762 1.0825 9.4693 4.320 0.00172 **
conditionface -2.6422 0.3316 1486.0508 -7.969 3.15e-15 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr)
conditionfc -0.152
We see that there is a highly (statistically) significant reduction of N170 voltages for faces compared to cars, but also that the estimate and \(p\)-value are slightly different compared to the previous models (based on the averaged data, with all the drawbacks mentioned above).
Note that there are other methods for statistical analysis of ERP data, such as cluster-based permutation tests (CBPTs) (Maris & Oostenveld, 2007, Sassenhagen & Draschkow, 2019). These do not require a strict a priori hypothesis about the time window and channel(s) of interest, and are therefore especially useful for exploratory analyses.
A tutorial for how to compute CBPTs in the MNE-Python and hu-neuro-pipeline packages will be included as a bonus chapter in the future.
6.6. Exercises#
Re-run the analysis pipeline for 10 participants from a different ERP CORE experiment (valid experiment names are
'N170','MMN','N2pc','N400','P3', or'ERN'), average the single trial amplitudes of the component of interest for each participant and condition, and fit a \(t\)-test to these averaged amplitudes.Repeat the same analysis but with the single trial data and a linear mixed model. Try to see if you can include random intercepts and slopes for participants, and random intercepts or slopes for items (if appropriate). Simplify the random effects in case the model fails to converge, and try to interpret the fixed effect estimates and \(p\)-values.
# Your code goes here
...
6.7. Further reading#
Paper Group-level EEG-processing pipeline for flexible single trial-based analyses including linear mixed models (Frömer et al, 2018)
Chapter Principles of statistical analyses: Old and new tools (Kretzschmar & Alday, 2023)
Blog post by Benedikt Ehinger on why to (almost) always include random slopes in LMMs
6.8. References#
- Bates et al., 2015
Bates, D., Mächler, M., Bolker, B., & Walker, S. (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67(1), 1–48. doi:10.18637/jss.v067.i01
- Burki et al., 2018
Bürki, A., Frossard, J., & Renaud, O. (2018). Accounting for stimulus and participant effects in event-related potential analyses to increase the replicability of studies. Journal of Neuroscience Methods, 309, 218–227. doi:10.1016/j.jneumeth.2018.09.016
- Fromer et al., 2018(1,2)
Frömer, R., Maier, M., & Abdel Rahman, R. (2018). Group-level EEG-processing pipeline for flexible single trial-based analyses including linear mixed models. Frontiers in Neuroscience, 12, 48. doi:10.3389/fnins.2018.00048
- Judd et al., 2012
Judd, C. M., Westfall, J., & Kenny, D. A. (2012). Treating stimuli as a random factor in social psychology: A new and comprehensive solution to a pervasive but largely ignored problem. Journal of Personality and Social Psychology, 103(1), 54–69. doi:10.1037/a0028347
- Kretzschmar & Alday, 2023
Kretzschmar, F., & Alday, P. M. (2023). Grimaldi, M., Brattico, E., & Shtyrov, Y. (Eds.). Principles of statistical analyses: Old and new tools. Language Electrified. Techniques, Methods, Applications, and Future Perspectives in the Neurophysiological Investigation of Language. Humana. https://doi.org/10.31234/osf.io/nyj3k.
- Kuznetsova et al., 2017
Kuznetsova, A., Brockhoff, P. B., & Christensen, R. H. B. (2017). lmerTest package: Tests in linear mixed effects models. Journal of statistical software, 82(13), 1–26. doi:10.18637/jss.v082.i13
- Maris & Oostenveld, 2007
Maris, E., & Oostenveld, R. (2007). Nonparametric statistical testing of EEG- and MEG-data. Journal of Neuroscience Methods, 164(1), 177–190. doi:10.1016/j.jneumeth.2007.03.024
- Sassenhagen & Draschkow, 2019
Sassenhagen, J., & Draschkow, D. (2019). Cluster-based permutation tests of MEG/EEG data do not establish significance of effect latency or location. Psychophysiology, 56(6), e13335. URL: https://onlinelibrary.wiley.com/doi/abs/10.1111/psyp.13335, doi:10.1111/psyp.13335
- Volpert-Esmond et al., 2021
Volpert-Esmond, H. I., Page-Gould, E., & Bartholow, B. D. (2021). Using multilevel models for the analysis of event-related potentials. International Journal of Psychophysiology, 162, 145–156. doi:10.1016/j.ijpsycho.2021.02.006


